To access and download PowerPoint presentation on 'DNA replication' click on the link below:
Download slides
DNA Replication
• A
basic property of genetic material (DNA) is to replicate in a precise way so
that the genetic information can be transmitted from each cell to its progeny.
• DNA
Replication is a biological process that occurs in all living organisms and
copies their exact DNA. It involves the synthesis of daughter DNA molecules from
parent DNA molecule by the aid of DNA dependent DNA polymerase enzyme.
• It
is the basis for biological inheritance.
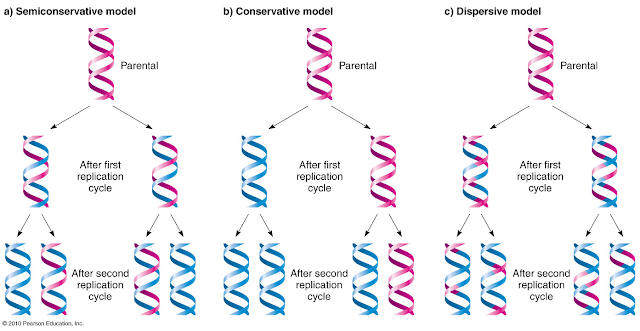
Possible models for DNA replication
• Conservative model: The parental molecule directs the synthesis of an
entirely new double-stranded molecule. Hence, after one round of replication,
one molecule is conserved as two old strands. This is repeated in the second
round.
• Semi-conservative model: The two parental strands separate and each serves as a
template to synthesize its complementary new strand. After one round of
replication, the two daughter molecules each comprise one old and one new
strand. After two rounds, two of the DNA molecules consist only of new
material, while the other two contain one old and one new strand.
• Dispersive model: Material in the two parental strands is distributed
more or less randomly between two daughter molecules.
DNA-dependent DNA polymerase
• DNA
replication is achieved by the enzyme known as DNA-dependent DNA polymerase
which requires single-stranded DNA as template.
• E.
coli
has 5 types of DNA polymerases:
– DNA
polymerase I: The first to be discovered. Required for excising the primers and
filling the gap during replication
– DNA
polymerase II: Functions in DNA repair machinery
– DNA
polymerase III: Main enzyme for replication
– DNA
polymerase IV: Functions in DNA repair machinery
– DNA
polymerase V: Functions in DNA repair machinery
• DNA
polymerase was first discovered by Arthur Kornberg and his colleagues in
1955.
•
Arthur Kornberg was awarded Nobel
Prize in Physiology or Medicine in 1959 for his discovery of the mechanism of
biological synthesis of DNA
Kornberg’s
discovery
• Kornberg
was trying to identify all the ingredients required to synthesize E. coli DNA
in vitro.
• First
successful DNA synthesis occurred in a mixture containing deoxyribonucleoside
5’-triphosphate precursors (dATP, dCTP, dTTP and dGTP, collectively called
dNTPs) and E. coli cell lysate.
• Kornberg
analyzed the mixture and isolated an enzyme that was capable of synthesizing
DNA.
• This
enzyme was originally called Kornberg enzyme but was later named as DNA
polymerase I.
• Subsequently
it was identified that DNA synthesis would not occur without the following 4
components:
– All
four dNTPs
– A
fragment of DNA as template
– DNA
polymerase
– Magnesium
ions
General features of DNA Polymerase
• Catalyze the polymerisation of deoxyribonucleotide precursors (dNTPs) into a DNA chain.
• At
the growing end of the DNA chain, DNA polymerase catalyzes the formation of a
phosphodiester bond between the 3’-OH group of the deoxyribose of the last
nucleotide with the 5’-PO4 group of the dNTP precursor.
• At
each step, DNA polymerase finds the correct precursor dNTP that is
complementary to the nucleotide on the template strand.
• The
process doesn’t occur with 100% accuracy, but error frequency is very low (10-6).
• DNA
synthesis always occurs in 5’ to 3’ direction.
• DNA
polymerase requires RNA primer to initiate DNA synthesis which provides a 3’-OH
group.
Process of DNA synthesis
• The
entire process of DNA synthesis can be broadly divided into three steps:
– Initiation
– Elongation
– Termination
A. Initiation
- The initiation of replication is directed by a DNA sequence called replicator which includes the origin of replication.
- Replication of E. coli chromosome is initiated at a 245 bp replication origin called oriC locus consisting of two series of short repeats:
o three repeats of a 13bp AT rich sequence
o four repeats of a 9bp sequence
- There are 8 different enzymes that together form a pre-priming complex for the subsequent reactions.
- A Dna A protein (initiating protein) binds to the four
repeats of 9 bp sequence of oriC. With the help of histone-like HU
proteins and ATP, the Dna A causes the AT rich 13 bp repeat region to
denature.
- The hexameric Dna B protein (helicase) now binds to
this region with the help of Dna C protein.
- The helicase then unwinds the DNA molecule bidirectionally creating
a replication bubble with two replication forks. The energy for unwinding
is derived from ATP hydrolysis – a reaction that causes conformational
change in helicase enabling it to move along a single strand of DNA.
• Next, multiple SSB (single-strand binding) proteins
bind cooperatively to separate the single-stranded DNA and prevents
renaturation.
• Continued unwinding by helicase causes supercoiling
ahead of the replication fork which is relieved by gyrase (topoisomerase
II).
• Other proteins associated with the initiation complex
include Dna T, Dna J, Dna K, Pri A, Pri B and Pri C.
B. Elongation
- Elongation includes both leading and lagging strand synthesis always in the 5’ to 3’ direction.
- As the two DNA strands are anti-parallel, the template strand is read in the 3’ to 5’ direction.
- For simultaneous synthesis of both the strands the synthesis of one strand is continuous while that of the other strand is discontinuous. Hence, DNA synthesis, as a whole, is semi-discontinuous.
- Each replication bubble has two replication forks moving in opposite directions.
- The anti-parallel nature of DNA strands allows both the strands to serve as templates and both strands of DNA are synthesized simultaneously.
- Therefore, DNA synthesis occurs simultaneously in both directions. The process of replication is, thus, bidirectional.
- The strand which is synthesized continuously in the same direction of the replication fork movement is called the leading strand.
- The discontinuous strand whose synthesis proceeds in the opposite direction of that of the replication fork movement is called the lagging strand.
- Leading strand synthesis begins with the formation of a short RNA primer at the replication origin. This is required because DNA polymerase cannot start de novo DNA synthesis but required a 3’-OH group to begin polymerization. The RNA primer is synthesized by primase (Dna G).
- The dNTPs are then added one by one to this primer by DNA polymerase III.
- Strand elongation occurs continuously keeping pace with the replication fork movement whose synthesis and positioning is done by an assembly of proteins called primosome. Primosome includes a helicase and a primase, along with five other proteins in E. coli.
• The short stretches of DNA in the lagging strand are
called Okazaki fragments after the name of its discoverer Reiji Okazaki.
• A primimg event is required to initiate each Okazaki
fragment during the discontinuous synthesis of the lagging strand.
• After each round of fragment synthesis, the RNA primer
is removed by the 5’ to 3’ exonuclease activity of DNA polymerase I and
the gap is filled with DNA by the polymerase activity of the same enzyme. The
remaining nicks are sealed by DNA ligase.
C. Termination
- Ultimately the two replication forks meet at the other side of the
circular DNA of E. coli.
- The terminus is a large region flanked by seven terminator sites:
- Ter E, Ter D and Ter A on one side
- Ter G, Ter F, Ter B and Ter C on the other side
- These terminator sites act as one-way valves that allow replication
forks to enter the terminus region but not to leave it.
- Tus protein arrests the movement of the replication fork at Ter sites. It
prevents unwinding of the helix by helicase.
- The final step is the topological unlinking of the two replication
products by topoisomerase II.
Proteins required for replication
• DNA
polymerase III: Required for dNTP polymerization and
proof-reading
• DNA
helicase: Unwinds the helix
• Primase:
Synthesizes
RNA primer
• Single
strand binding proteins: Prevents renaturation of the
replication bubble
• Topoisomerase
II: Relieves
supercoiling ahead of the replication fork as well as causes topological
unlinking of the two replication products at termination.
• RNase H: Removes RNA primer
• DNA
polymerase I: Fills the gap after removal of RNA
primers with DNA and removes errors
• DNA
ligase: Seals the single-stranded nick in DNA
• DNA sliding clamp: slides along the DNA template and keeps the polymerase
from falling off the template.
DNA helicase
• A class of enzymes that couples ATP hydrolysis with the unwinding of DNA double helix.
• They are typically hexameric proteins in the shape of
a ring.
SSB proteins
- As the DNA strands separate, the SSB proteins bind co-operatively to the single-stranded DNA.
- They prevent the renaturation of the unwound DNA strands.
Topoisomerases
• As the DNA unwinds, the DNA strands get positively
supercoiled ahead of the replication fork.
• This supercoiling is relieved by topoisomerases
(Gyrase in E. coli). The energy for such action is obtained from ATP
hydrolysis.
Primase
- Primase is a specialized RNA polymerase which use ssDNA as a template to synthesize short RNA primers.
- It is activated by interacting with DNA helicase.
DNA polymerase
• DNA synthesis is catalyzed by the enzyme DNA
polymerase (DNA polymerase III in E. coli).
• Using an ssDNA as a template, it can catalyze the formation
of a phosphodiester bond between a 3’-OH of the primer and 5’-phosphate of the
incoming dNTP.
• It cannot tart DNA synthesis de novo.
Sliding DNA clamps
• Surrounds the DNA and binds the polymerase.
• Slides along the DNA template and keeps the polymerase
from falling off the template.
• Increases the processivity of the polymerase.
Rnase H
- The RNA primers are removed by RNase H.
- H stands for ‘hybrid’ since they degrade RNA from the RNA-DNA hybrids.
DNA polymerase
• Removal of the RNA primers leaves single-stranded gaps
in the DNA.
• These gaps are filled by DNA polymerase (DNA
polymerase I in E. coli)
DNA ligase
- DNA ligase seals the nick.
- It catalyzes phosphodiester bond formation using energy derived from ATP hydrolysis.
DNA Polymerase I
• Encoded
by polA gene.
• Consists
of a single polypeptide of 109 kDa.
• Possess
following catalytic activities:
– 5’ → 3’ polymerase activity
– 3’ → 5’ exonuclease activity
– 5’ → 3’ exonuclease activity
• Involved in proof-reading of replicated DNA by
3’ → 5’ exonuclease activity.
• Involved
in repairing damaged DNA by unique 5’ → 3’ exonuclease activity as well as
removing RNA primers and filling the gaps.
DNA Polymerase III
• A
multimeric enzyme with molecular mass 900 kDa
• Minimal
core (has catalytic activity in vitro) contains 3
subunits: α,
ε
and θ.
• Catalytic
core synthesizes short DNA fragments and falls off the template
• The
τ subunit results in dimerization of the catalytic core and
increased activity.
• The
β
subunit forms a dimeric clamp, keeps the catalytic
core from falling off the template and slides along it.
• A
group of 5 other subunits (γ,
δ,
δ’,
χ
and ψ)
forms the γ
complex which loads the enzyme onto the template at
the replication fork.
• Holoenzyme
possesses 5’ → 3’ polymerase activity and 3’ → 5’ exonuclease (proof
reading) activity.
Processivity
• The ability of an enzyme to catalyze many reactions
before releasing its substrate is called processivity.
• The sliding clamp loader loads the sliding clamp to
DNA polymerase and increases its processivity.
• DNA polymerase can add upto 1000 nucleotides / second
to the growing nucleotide chain.





Very informative. Thank you ��
ReplyDelete